BOED in Enzyme Kinetics
Apply Bayesian optimal experimental design (BOED) to biocatalysis problems. Cooperation with Jürgen Pleiss and Jan Range.
This Project is a joint work of Jürgen Pleiss and Jan Range from the Institute of Biochemistry and Technical Biochemistry and Nicole Radde and Sebastian Höpfl from the Institute for Systems Theory and Automatic Control.
This project aims to enable more efficient experiment planning by predicting the best times for an experiment and thus avoiding costs for many iterations.
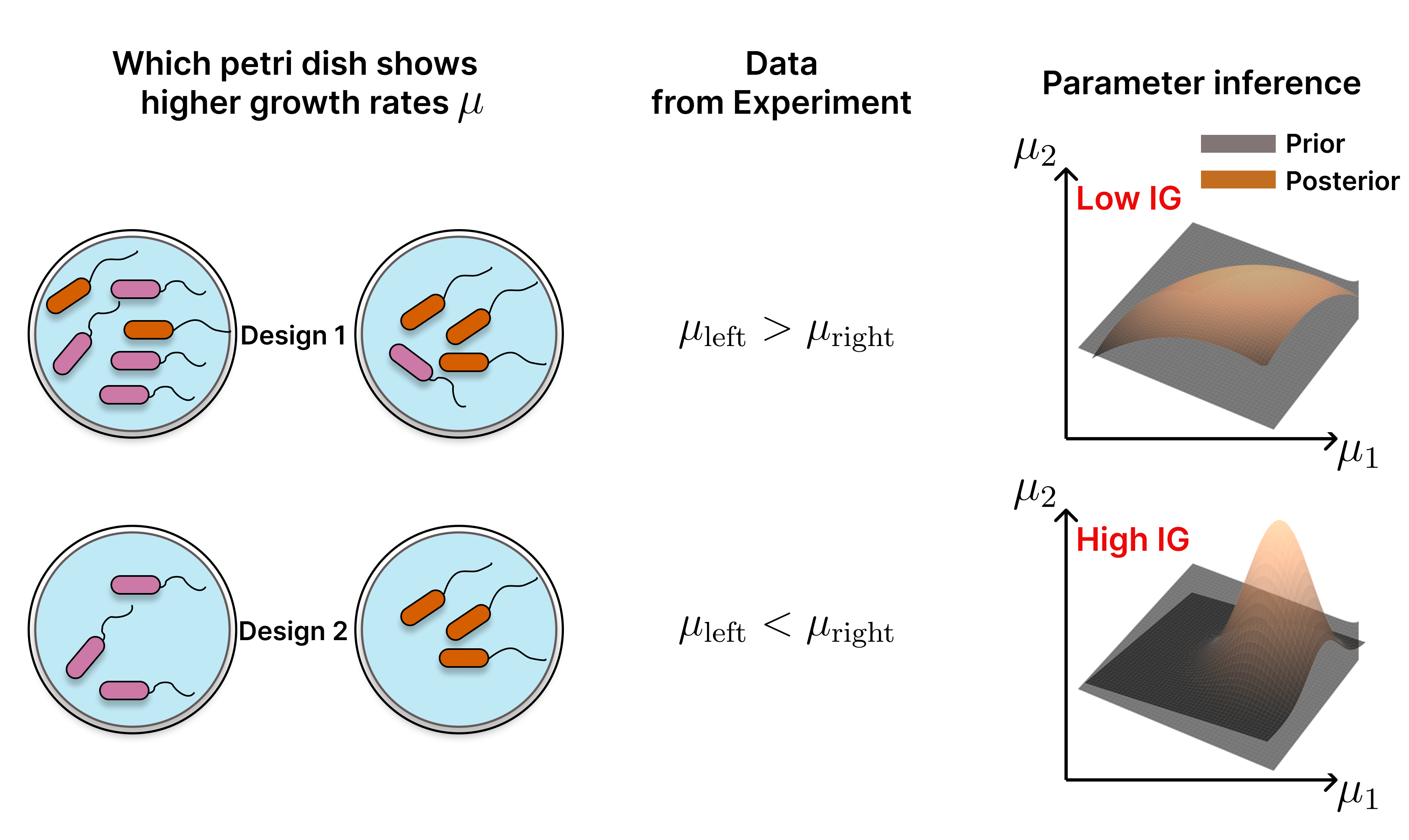
Bayesian Optimal Experiment Design (BOED) favours the experiment, which maximizes the Information Gain (IG) in the parameter estimation. Here we select between two Designs wich measure the growth rate $\mu$ in two petri dishes. The outcome of the experiment is which petri dish shows higher overall growth rates and the value of these apparent growth rates. The second Design is more informative as the growth rates of both bacteria species can be determined separately. Therefore the posterior of Design 2 is more informative as in Design 1 and both start with a flat prior.
For this project we use EnzymeML and PEtab to exchange parameter estimation problems in a FAIR manner.
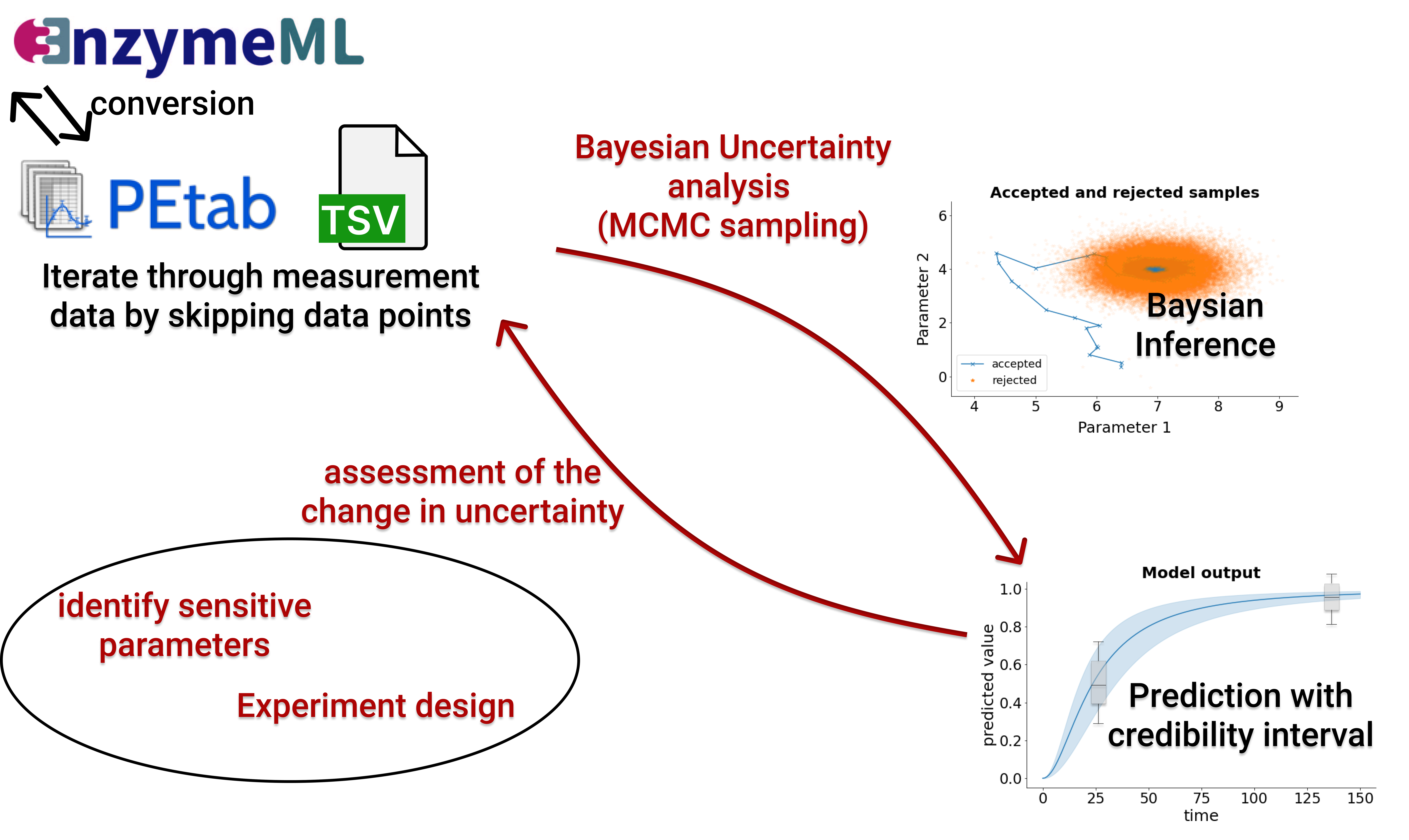
The EnzymeML and PEtab formats can be converted to each other. PEtab is used to perform the bayesian uncertainty analysis via MCMC or Emcee sampling. The results of this analysis are credibility intervalls of the estimated parameters. These results are then stored in the according PEtab and EnzymeML file again.