QuaLiPerF
Statistical methods for data integration into multi-scale models and uncertainty tracking
QuaLiPerF Quantifying Liver Perfusion-Function Relationship in Complex Resection – A Systems Medicine Approach
As Doctoral Researcher in the DFG Research Unit QuaLiPerF, I am working towards an advanced holistic understanding of liver function in the context of liver resection. The liver is the body’s most important metabolic organ. It is responsible for detoxification, as well as the storage of vitamins, glycogen and iron. Toxins like drugs, hormones or ammonia are excreted through the intestine via the biliary fluid or through the kidneys via urine.
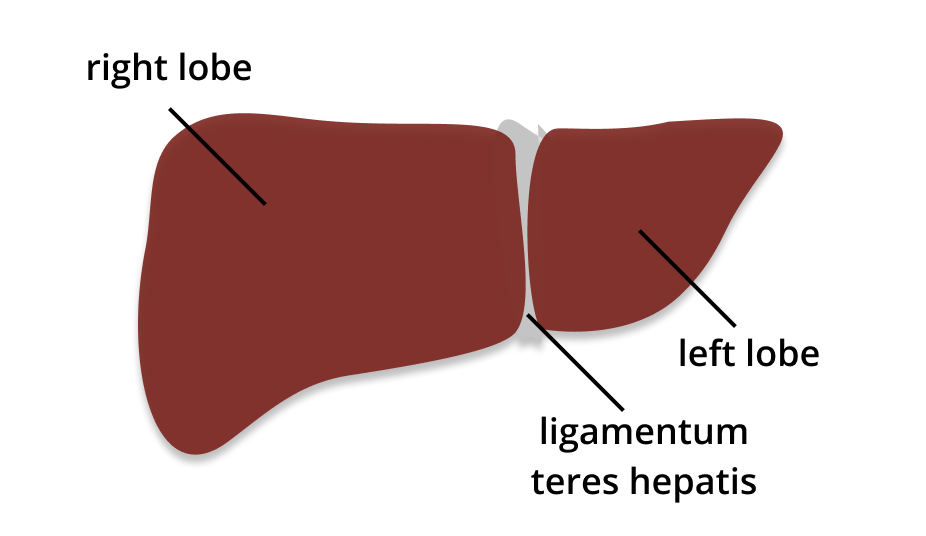
The need for extended hepatectomy due to tumours is rising in our aging society, while the risk for liver failure after resection is still high for patients with pre-existing hepatic diseases like fibrosis, hepatitis or steatosis. Today two-stage partial hepatectomy approaches allow liver resection of 70 %, where the liver remnant can recover its original size completely.
As the liver is a highly complex organ, functional capacity is not evenly distributed. Therefore, consideration of only the remnant liver size is not sufficient to be able to accurately assess the functionality of the liver remnant (Sparrelid et al. 2017). However, a good guess for the remaining liver function is essential to prevent liver failure, especially in extended liver resections. Thereby, surgeons must find a good compromise between removing enough tissue to prevent reoccurrence of the tumour and leaving enough tissue to ensure that the liver remnant has enough functional capacity left (Christ et al. 2017). To assess the functional liver capacity in the context of hepatectomy, QuaLiPerF combines metabolic profiling and liver metabolism on the cellular scale with hepatic perfusion and functional distribution on the lobular scale. This goal is reached through the combination of experimental and modelling approaches.
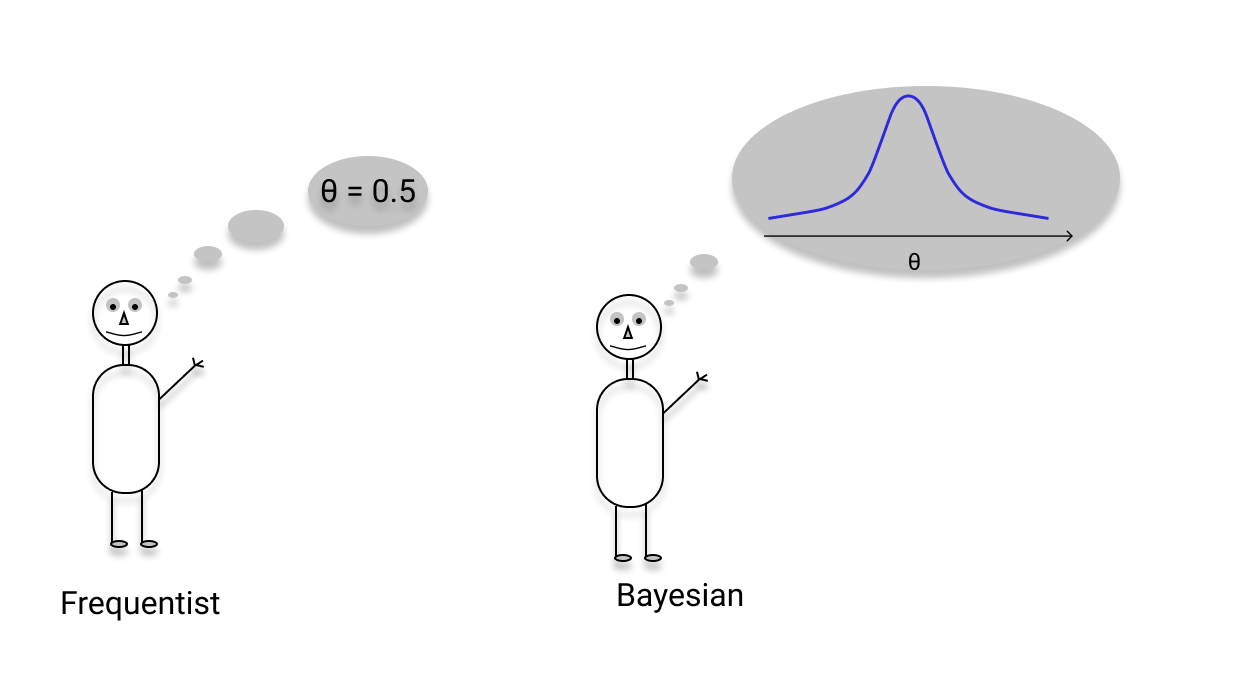
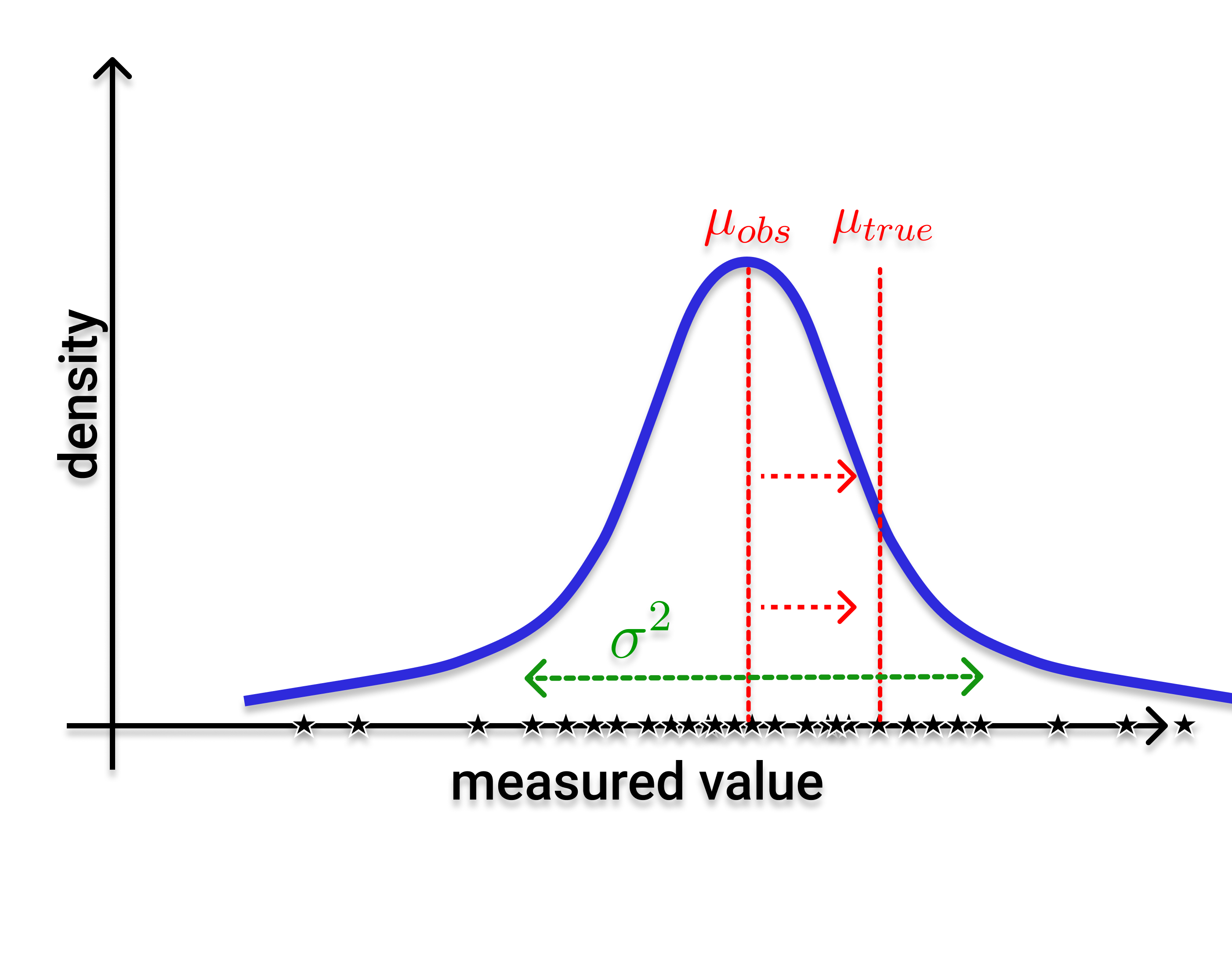
My focus in the research unit QuaLiPerF is to apply statistical methods for data integration into multi-scale models and uncertainty tracking (P5). This is done in cooperation with the modelling and experimental project partners, providing a stochastic way to track uncertainty in models on the cellular, lobular and later whole-body scale. Therefore, this project forms a framework around the other model projects of the research unit and should in the end yield to coupled models on multiple scales.
To achieve this, statistical methods like Markov chain Monte Carlo (MCMC) sampling, Maximum Likelihood (ML) parameter estimation and machine learning will be applied. For example, by defining error functions for ODE models, making the optimization of a likelihood optimization problem feasible and allowing to transfer uncertainties in the data to uncertainties in the model output. Machine learning approaches could help to smartly select high impact parameters for individualized assessment of regeneration capacity and assessing the risk of liver failure.
A further aim is to translate the models based on rodent data to models that can be used for patients in a systems medicine approach. This should ultimately lead to individualized models for human patients in the context of liver hepatectomy.
References:
Christ, Bruno; Dahmen, Uta; Herrmann, Karl-Heinz; König, Matthias; Reichenbach, Jürgen R.; Ricken, Tim et al. (2017): Computational Modeling in Liver Surgery. In: Frontiers in physiology 8, S. 906. DOI: 10.3389/fphys.2017.00906.
Sparrelid, Ernesto; Jonas, Eduard; Tzortzakakis, Antonios; Dahlén, Ulrika; Murquist, Gustav; Brismar, Torkel et al. (2017): Dynamic Evaluation of Liver Volume and Function in Associating Liver Partition and Portal Vein Ligation for Staged Hepatectomy. In: Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract 21 (6), S. 967–974. DOI: 10.1007/s11605-017-3389-y.